Parallel Genomics Data Processing
Published
With sequencing costs falling rapidly, the demand for production grade, parallel genomics data processing is increasing.
Executive Summary
With the ability to perform Whole Genome Sequencing becoming cheaper than ever, we can expect to see an explosion in the amount of data generated by sequencers.
At present, the toolchains for processing this raw data into valuable insights are rooted in academia, and unintended for high throughput processing. The only production toolchain on the market is Nvidia Clara Parabricks, a tiny segment of their business with high licensing fees.
A startup providing a well engineered, GPU-based cloud pipeline could capitalize on this new influx of data.
Era of low cost, ultra high throughput sequencing
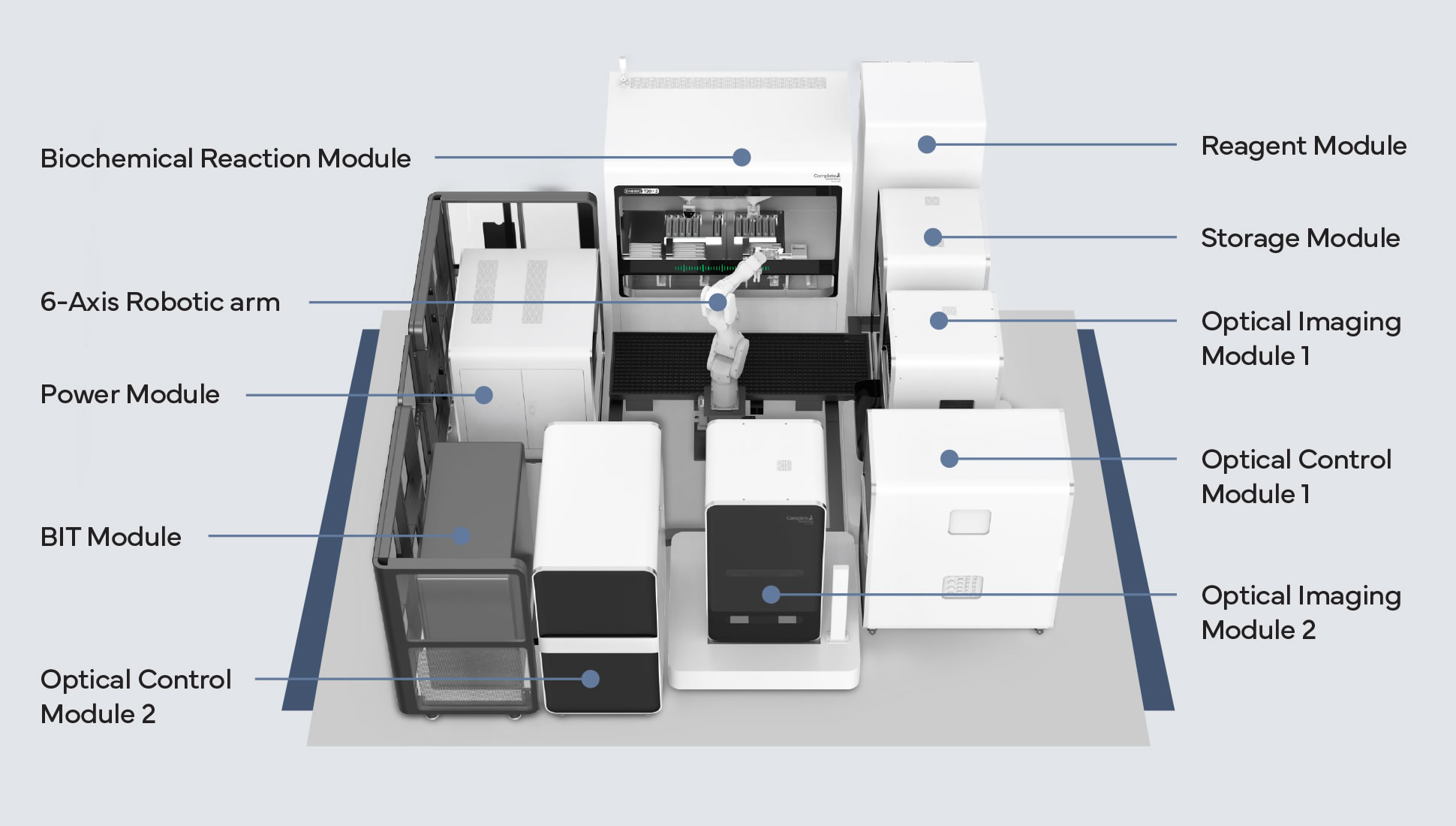
The sequencer above, the BGI T20x2, is a new breed of ultra high throughput sequencing device, capable of producing in excess of 140Tb of genomic data per week. The volume of data production is vastly outpacing the computational infrastructure that labs currently have. Their current solution for this is to be choosy with which genomes they sequence and why.
AI scale datasets
In order to train models capable of untangling gene functions across populations, we will require datasets on the order of 100M genomes. The current largest academic dataset of genomes, the UK Biobank has at approximately 500k genomes.
